ProjectABSTRACTMETHODOLOGY WORK PLAN |
RESEARCH METHODOLOGY
Within 2 years, samples will be collected seasonally in the Gdańsk Deep and in 4 additional stations (Gotland Deep, Bornholm Deep and 2 in the Gotland Basin) (Fig.1) in 2 seasons (spring and autumn) in order to have representative spatial and temporal coverage. Cruises will be arranged on the r/v Oceania. 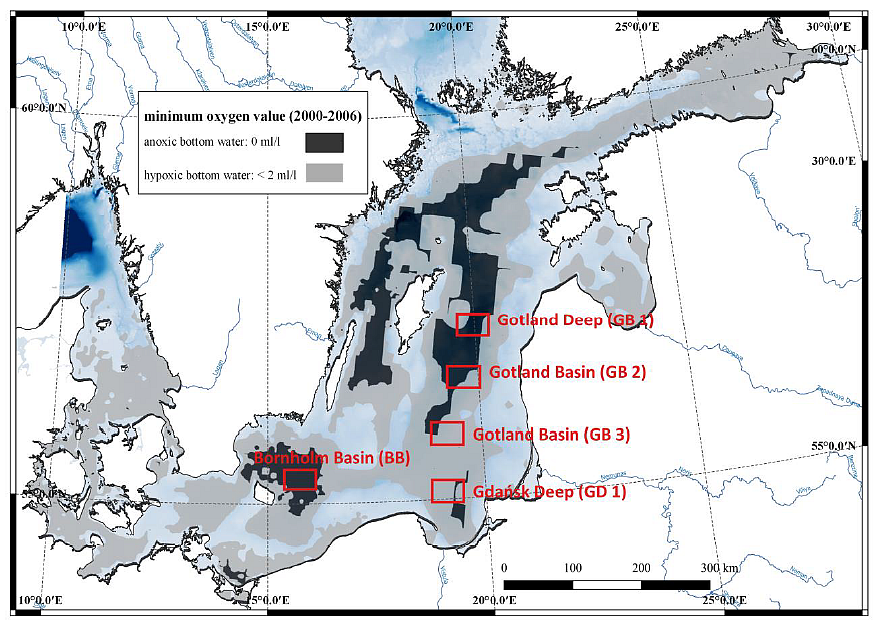
Fig. 1. Sampling sites and the oxygen concentration in the Baltic Sea bottom water in 2000-2006. GIS oxygen datasets produced by the BALANCE project. Sea water samples will be collected by CTD rosette sampler with 10 Niskin bottles while sediment samples will be collected by means of the Niemisto gravity corer. Pore water will be collected by means of Rhizones. Samples for incubation experiments, metagenomic and metatranscriptomic analyse and auxiliary parameters will be collected from the oxygen minimum zones and sediments. The sampling strategy is presented in Fig. 2. Procedures of water and sediment incubation experiments will be modified after Dalsgaard et al. (2003, 2005, 2013), Deutsch et al. (2010) and Bonaglia et al. (2014, 2016). All samples from incubations experiments for N2/Ar ratio (and nitrogen isotopes) will be measured via membrane-inlet mass spectrometry (MIMS), BAY INSTRUMENTS (Kana et al. 1994). The quality control will be performed always before implementing the method. 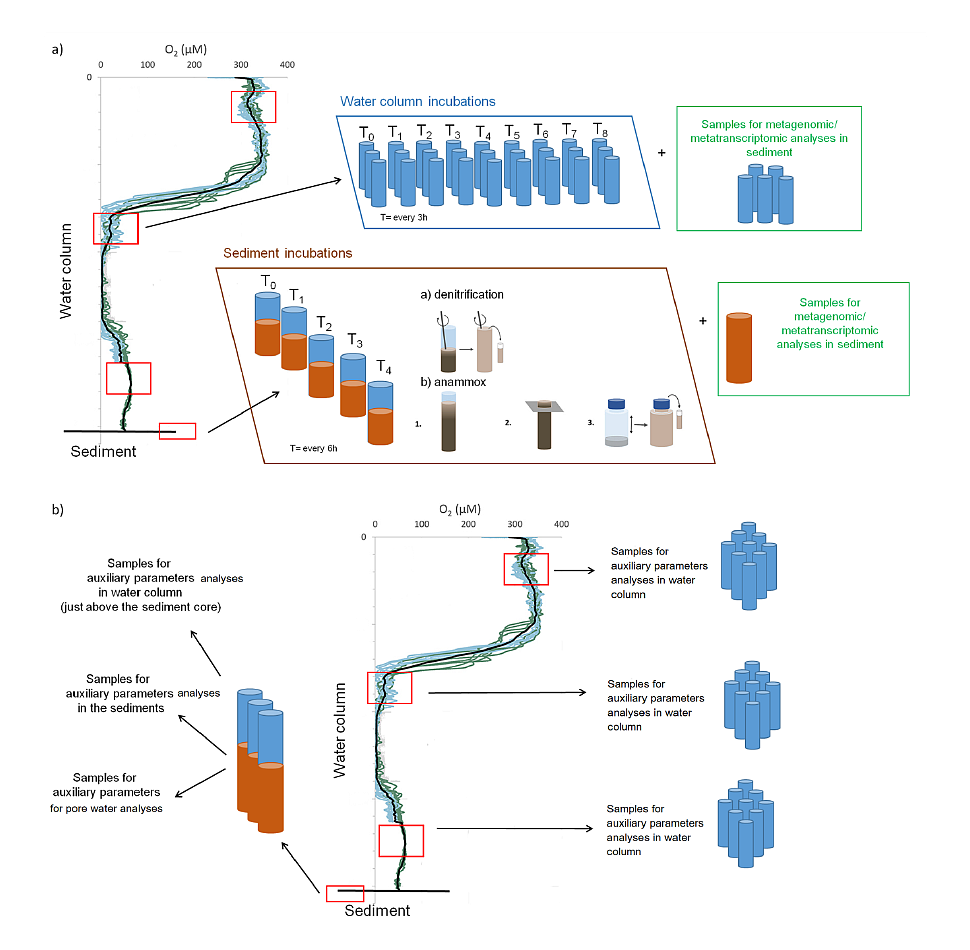
Fig. 2. Scheme of the sampling strategy including a) collecting samples for performing the incubations experiments and metagenomic/metatranscriptomic analyses and b) collecting samples for of auxiliary parameters analyses. The presence of bacterial DNA in the collected samples and the lack of inhibitors will be confirmed by positive PCR reactions with primers F27 and 1492 complementary to the 16S rRNA gene. In each sample, using DNA template qPCR will be used to quantify an abundance of N-cycle genes especially those possibly active in Baltic Sea hypoxia: (e,g,: napa, narG, nirK, nirS, nrfA, nor, nosZ, nod, hzsA). Additionally, metagenomic and metatranscriptomic analysis will be used for monitoring microbial community structure and functioning. This project intends to deposit the information about obtained DNA sequences in public sequence databases (e.g. NCBI, RDP). All NGS sequencing will be outsourced to a specialized external company, selected based on the current market situation. The auxiliary parameters (Table 1) analyses are regularly done in the Host Institution (IO PAN) and will be performed with the state of the art methods, while the quality control will be checked by certificate standards and reference materials. For the purpose of the proposed project, the standard method for nutrients analyses cannot be used due to both too high detection limits and too big volume of sample needed for analyses. Nutrients always will be simultaneously measured to N2/Ar and there will be not enough volume of sample for each analysis. Therefore within this project nutrient measures will be performed by means of the globally recommended Continuous Flow Analyzer. The sophisticated statistical analyses such as e.g. Principal Components Analysis (PCA), principal component regression (PCR), ANOVA will be used by STATISTICA software. Table 1. List of the auxiliary parameters measured in designated samples
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||


